Main objectives of the project include:
1. Genetic susceptibility in AD
Identify and interpret common and rare variants contributing to AD risk across diverse cohorts and ancestries.
2. From GWAS loci to causal genes
Fine-map AD GWAS signals, prioritize coding and regulatory variants in linkage disequilibrium with known markers, and define the most likely causal genes.
3. Shared genetic architecture with synucleinopathies
Quantify genetic overlap and divergence between AD, PD, DLB, RBD, and MSA to uncover convergent pathways and disease-specific mechanisms.
4. Genotype–phenotype correlations
Study how genetic profiles relate to age of onset, progression, cognitive trajectories, imaging biomarkers, and clinical subtypes.
5. Epigenetic and regulatory mechanisms
Investigate AD-related changes in DNA methylation, histone modifications, chromatin accessibility, and non-coding RNA expression, with emphasis on brain cell-type specificity.
6. Functional follow-up in cellular models
Prioritize high-confidence genes for experimental validation in patient-derived cellular systems (e.g., iPSC-derived neurons/microglia) and other model platforms.
Our AD genetics project includes multiple sub-projects, for example:
- In-house GWAS/WGS to identify and refine AD risk loci
- Genome-wide association meta-analyses
- Rare-variant burden and SKAT/SKAT-O analyses
- Multi-omics fine-mapping of novel loci
- Cell-type-specific regulatory annotation
- Polygenic risk score modeling
- Machine-learning pipelines for gene prioritization
- Systems biology and network crosstalk analysis
- Drug-target prioritization and repurposing
- Integration of WES/WGS datasets (ADNI, ROSMAP, ADSP, UKBB, TRIAD, PREVENT-AD, CIMA-Q, etc.)
Ultimately, our goal is to deliver a clearer map of the genetic and molecular landscape of AD, and to translate that knowledge into clinically meaningful pathways and therapeutic opportunities.
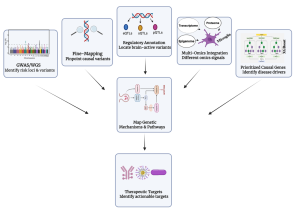
From GWAS to Causal Mechanisms in Alzheimer’s Disease
Integrative framework used in our lab to prioritize causal AD genes and identify actionable pathways by combining GWAS, rare-variant signals, and brain-specific multi-omics.